Computational Biophysics in COMSOL®: FSI-Simulations of Cells in a Microfluidic Device
INTRODUCTION
The mechanical properties of biological cells are promising biomarkers to differentiate for example cell phenotypes, cell states or between healthy and unhealthy cells with applications ranging from research facilities to medical laboratories [1, 2]. Real-time deformability cytometry allows probing the mechanical characteristics of ~1000 cells / s by imaging the cells flowing through a microfluidic channel [1]. The observed deformation can be used to infer the mechanical properties. So far, the expected deformation has been analysed theoretically in [3] and numerically in [4] assuming only a homogeneous elastic material of the cell. In this project, we extend the numerical model to incorporate viscosity. Due to the time-dependent response of a viscoelastic material, we have to extend the framework to take into account the entry and the exit of the cell in the narrow channel respectively out of the channel.
USE OF COMSOL MULTIPHYSICS® SOFTWARE
We use COMSOL Multiphysics® to perform the two-way coupled FSI simulations in a 2D axisymmetric and 3D geometry. The Laminar Flow interface controls the fluid flow in the microfluidic channel. A Solid Mechanics interface defines the viscoelastic properties of the biological cell. The Multiphysics Branch accomplishes the two-way FSI interaction between the fluid and the solid sphere. We move the cell through the fluid channel by the Moving Mesh interface eliminating the need of remeshing.
RESULTS
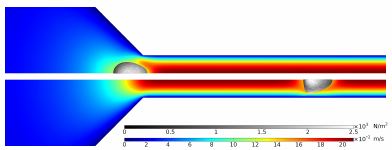
So far, we managed to reproduce the results of [4] in COMSOL and are currently extending the model to take into account the viscosity of the cell. In [4], the flow field is artificially modified to keep the cell stationary in the flow channel which permits meaningful results for viscoelastic cells. Here, we present an extended simulation where we move the mesh surrounding the cell with the same speed as the cell through the channel such that we do not even have to remesh (see Figure 1).
CONCLUSION
With this numerical setup, we will be able to analyse the impact of the entrance and exit of the narrow flow channel to the viscoelastic cell. Those insights will then be incorporated into RT-DC to enable simultaneous extraction of cell elasticity and viscosity.
REFERENCES
- Otto et al., Real-time deformability cytometry: on-the-fly cell mechanical phenotyping, Nat. Methods, 2015
- Henry et al., "Quantitative diagnosis of malignant pleural effusions by single-cell mechanophenotyping", Sci. Transl. Med., 2013
- Mietke et al., "Extracting Cell Stiffness from Real-Time Deformability Cytometry: Theory and Experiment", Biophys. J., 2015
- Mokbel et al., "Numerical Simulation of Real-Time Deformability Cytometry To Extract Cell Mechanical Properties", ACS Biomater. Sci. Eng., 2017
Download
- wittwer_poster.pdf - 0.91MB
- wittwer_abstract.pdf - 0.11MB
